Product Description
Monkeypox Virus Real Time PCR Kit | YJC70115NW- 50 | BioPerfectus
1. Intended Use
The Bioperfectus Monkeypox Virus Real Time PCR Kit is an in vitro diagnostic test, based on real-time PCR technology, for the detection of DNA from the Monkeypox virus. Specimens can be obtained from human tonsillar swab, nasopharyngeal swab, serum, whole blood, lesion exudate and scab. BSL-2 facilities with standard BSL-2 work practices may be used for the test of the Monkeypox virus.
2. Kit Components

3. Storage
- All reagents should be stored at -20±5°C condition.
- Check expiry date before use and do not use expired reagent.
- Keep detection mix away from light.
- Avoid repeatedly freeze-thaw.
- Manufacturing date and expiry date: see outer packing box. 4. Materials and Devices Required but Not Provided.
- Appropriate real time PCR instrument: Bioperfectus STC-96A, STC-96A PLUS, Applied Biosystems 7500, QuantStudio™ 5, Roche LightCycler®480, Bio-Rad CFX96™, QIAGEN Rotor-Gene Q, Analytik Jena qTOWER 3 and other applicable Bioperfectus machines.
- Appropriate Nucleic acid extractor:SSNP-2000B (32 channels), SSNP-3000A (64
channels), SSNP-9600A (96 channels), SMPE-960 (96 channels), SAW-96 (96 channels), SAW-48 (48 channels) and other applicable Bioperfectus machines. - Magnetic grate for 1.5 mL centrifuge tubes.
- Centrifuge tube shelf.
- Centrifuge with a rotor for 1.5 mL reaction tubes.
- Centrifuge with a rotor for 0.2 mL reaction tubes or plate.
- Vortex mixer.
- Calibrated adjustable pipettes or multi-channel pipette.
- Pipette tips with filters.
- 1.5mL centrifuge tubes.
- 0.2 mL PCR tubes or plates.
- Disposable particle-free gloves and operating grown.
- 10% sodium hypochlorite or pasteurized disinfectant.
- Biological safety cabinet or PCR hood.
5. Background Information
Monkeypox is a rare disease that is caused by infection with monkeypox virus. Monkeypox virus belongs to the Orthopoxvirus genus in the family Poxviridae. The Orthopoxvirus genus also includes variola virus (which causes smallpox), vaccinia virus (used in the smallpox vaccine), and cowpox virus. Monkeypox was first discovered in 1958 when two outbreaks of a pox-like disease occurred in colonies of monkeys kept for research, hence the name ‘monkeypox.’ In humans, the
symptoms of monkeypox are similar to but milder than the symptoms of smallpox. Monkeypox begins with fever, headache, muscle aches, and exhaustion. The main difference between symptoms of smallpox and monkeypox is that monkeypox causes lymph nodes to swell (lymphadenopathy) while smallpox does not. Transmission of monkeypox virus occurs when a person comes into contact with the virus from an animal, human, or materials contaminated with the virus. The virus enters the body through broken skin (even if not visible), respiratory tract, or
the mucous membranes (eyes, nose, or mouth). Laboratory tests that are used to diagnose monkeypox virus include detection of immunohistochemical testing, electron microscopy, real time polymerase chain reaction (RT-PCR), and virus isolation.
6. Technical Principle
The Bioperfectus Monkeypox Virus Real Time PCR Kit is based on real-time PCR technology. Specific primers and probes are designed based on F3L gene areas of Monkeypox virus. Probes consist of a reporter dye at 5’ and quenching dye at 3’. The fluorescent signals emitted from reporter dye are absorbed by the quencher, soit doesn’t emit signals. During amplification, probes bonded to templates are cut off by Taq enzyme (5’→3’ exonuclease activity), separating reporter dye fromthequencher, generating fluorescent signals, the PCR instrument will thenautomatically draw a real-time amplification curve based on the signal change, finally realizing the qualitative detection and differentiation of DNAfromMonkeypox virus. In addition, the kit also contains a housekeeping gene (RNaseP) as an internal control (IC) for specimen sampling and nucleic acid extraction.
7. Warnings and Precautions
- For in vitro diagnostic use only. For professional use only.
- Operators should be trained in real-time PCR techniques.
- All patient specimens should be inactivated at 56°C for 30 minutes andprocessed in accordance with laboratory biosafety requirements.
- Nucleic acid extraction should be manually carried out in biosafety cabinet or byautomatic nucleic acid extraction system.
- Wear personal protective equipment (PPE), including (but not limitedto)
disposable clean powder-free gloves, mask, goggles. - Working zones in laboratory should be strictly separated. Use separatedandsegregated working areas for (i) Reagent preparation, (ii) Specimen preparationand (iii) Amplification. The workflow in the laboratory should proceedinunidirectional manner. The experiment processes shall comply with the GoodClinical Laboratory Practice (GCLP) for Molecular Based Tests UsedinDiagnostic Laboratories.
- Work benches should be cleaned immediately after use. Ampliconcontamination should be avoided.
- Clean work benches, pipettes and centrifuge by using 10% sodiumhypochloriteand 70% ethanol.
- The use of sterile disposable pipettes and nuclease-free pipette tips isrecommended.
- Use applicable real-time PCR instrument and nucleic acid extraction systemtoensure optimal test performance.
- Use reagents before expiry date. DON’T replace or interchange reagents fromdifferent batches or manufactures.
- Discard specimens and assay waste according to your local safety regulations.
8. Sample Preparation
8.1 Sample collection method
For tonsillar swab:
- Swab or brush posterior tonsillar tissue with a sterile dry polyester swab.
- Break off end of applicator into a 1.5 or 2 mL screw-capped tube with O-ringor place entire swab in a sterile container.
For nasopharyngeal swab:
- Swab the nasopharynx with a sterile dry polyester swab.
- reak off end of applicator into a 1.5 or 2 mL screw-capped tube with O-ringor place entire swab in a sterile container.
For serum and whole blood:
- Collect 7 to 10 cc of patient blood into a red/gray (marbled), gold, or red
topped serum separator tube when patient is first identified. - Spin tubes to separate serum.
- Save the serum in at least 2 aliquots, 1 for immediate testing and the other for paired sera testing at the convalescent–stage of disease.
- Collect 3 to 5 cc of whole blood into a lavender-topped tube.
- Gently invert the tube to mix the blood with the anticoagulant.
- Obtain convalescent-phase serum 4 to 6 weeks after initial acute-phase serumcollection.
- Send convalescent-phase serum with the remaining acute-phase serumaliquot.
For lesion exudate:
- Sanitize lesion with an alcohol wipe, allow to dry.
- Use a disposable scalpel (or a sterile 26 Gauge needle) to open and remove thetop of the vesicle or pustule.
- Swab the base of the lesion with a sterile polyester or swab.
- Break off end of applicator into a 1.5 or 2 mL screw-capped tube with O-ringor place entire swab in a sterile container.
For scab:
- Sanitize skin with an alcohol wipe, allow to dry.
- Use a 26 Gauge needle to pick or dislodge at least 4 scabs; two scabs each fromat least two body locations.
- Place scabs from each location in separate sterile O-ring vials. Detailed instructions for specimen collection for cases of suspect monkeypox canbe found on the CDC website at https://www.cdc.gov/poxvirus/monkeypox/lab-personnel/lab-procedures.html 8.2 Transportation
Specimen packaging and transportation follows regulations. https://www.cdc.gov/smallpox/lab-personnel/specimen-collection/pack-transport.html - Pack 3 layers according to class A or B infectious articles if external transportation involves.
- Specimen collected from suspected cases should be preserved using 2-8°Cicebags or -70°C dry ice and sent to qualified laboratories within 24 hours.
8.3 Sample transport and storage
- Specimen preserves at 2-8°C up to 24 hours after received.
- Specimen preserves at -70°C or colder if extraction is arranged after 24 hours. Extracted DNA preserves at -70°C or colder.
9. Procedure
9.1 DNA Extraction
For swab, serum, whole blood and lesion exudate samples, transfer fluid samplesfor nucleic acid extraction according to the manufacturing instructions. For scab samples, mix 0.2g-1.0g of tissue samples with sterilized quartz-sandwithratio of 1:1 (v/v) and grind thoroughly, add PBS (0.01mol/L, pH 7.6-7.8) withratioof 1:5 (w/v) and freeze-thaw twice at -20℃, then centrifuge at 8,000 rpmfor 5minutes at 4℃, and transfer supernatant for nucleic acid extraction.
9.2 Master Mix Preparation
The Master Mix volume for each reaction should be pipetted as follows:
Determine the number of extracted specimens to be tested, thaw the components. For maximal recovery of contents, briefly spin vials in the centrifuge before opening. Mix carefully and thoroughly by pipetting up and down.
9.3 PCR Set-up
Procedure Place your samples on ice. Follow the procedure below to prepare the PCR Master Mix.
- a. Pipette 20μL of the Master Mix into each required reaction tubes/plate.
- b. Add 5μL isolated DNA or 5μL Controls (Positive Control or Blank Control).
- c. Make sure that every run including at least one Positive Control and one Blank Control.
- d. Cap or seal the reaction tubes/plate and centrifuge using an appropriate
centrifuge for 30 seconds at approximately 2,000 rpm. - e. Ensure that all liquid is at the bottom of the tubes/plate.
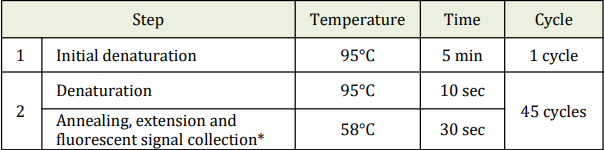
- f. Perform the following protocol in the instrument.
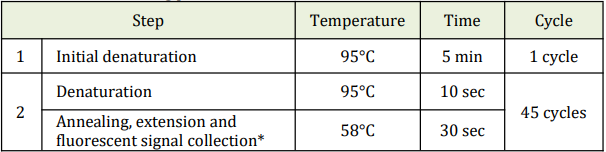
* Fluorescent signal should be collected during this step through the FAM and VIC
channels.
10. Real Time PCR System Operation.
The following amplification protocol was developed for use on the Bioperfectus STC-96A, STC-96A PLUS. See the instrument operator’s manual for detail. Other appropriate real time PCR instruments refer to the corresponding instrument operator’s manual.
10.1 Bioperfectus STC-96A/96A PLUS Real-Time PCR System Amplification Protocol
1. Switch on Bioperfectus STC-96A/96A PLUS Real-Time PCR System.
2. Launch the Bioperfectus STC-96A/96A PLUS Real-Time PCR System software Version 1.0.
3. Click on “Experiment Wizard”, and set up proper parameters in “Project” and "Setup”.
4. Set up "Plate”.
5. Set up “Sample”.
6. Starting the PCR
- a. Insert the 96 well PCR plate or reaction tubes into the machine.
- b. Select the “Start Run” button,
7. Post PCR Analyze the data by pressing the “Analysis” button on left side of the menu and analyze the data using the “Analyze”. 11. Quality Control Prior to evaluating the specimen results, the Positive Control and Blank Control should be interpreted using the table below.
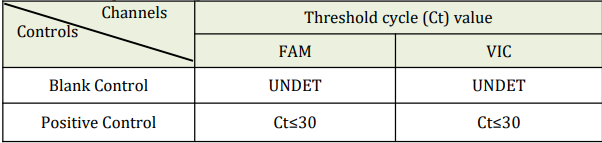
NOTE: Internal Control is specially designed to detect in fluorimeter channel VIC
- The Positive Control and Blank Control should be included per PCR run.
- If the Positive Control and Blank Control do not meet the criteria, the entire run
is invalid and results should not be reported. Repeat the entire process
(specimen and control preparation, amplification and detection). If the repeat run is still invalid, please contact Technical Support. - Viral transport media or previously characterized negative specimen may be used as an external negative control. This must be treated as a patient specimen in every extraction and PCR run.
- Additional controls may be used in accordance with local, state, federal accrediting organizations, as applicable.
12. Limitations
- Negative results do not preclude infection with Monkeypox virus and shouldnot be the sole basis of a patient treatment decision.
- Reliable results are dependent on the adequate specimen collection, transport, storage and processing procedures.
- Inhibitors present in the sample and/or errors in following the assay proceduremay lead to false negative results.
- A trained healthcare professional should interpret assay results in conjunctionwith the patient's medical history, clinical signs and symptoms, and the resultsof other diagnostic tests.
- Potential mutations within the target regions of the virus genome covered bythetests primers and/or probes may result in failure to detect the presence of thepathogens.
- There is a risk of false positive values resulting from cross-contaminationbytarget organisms, their nucleic acids or amplified product, or fromnon-specificsignals in the assay
13. Data Analysis and Interpretation
The following results are possible: FAM channel for Monkeypox virus, VIC channel
for IC.
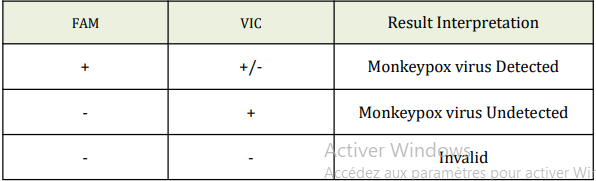
Note:For Monkeypox Virus: Ct value ≤40 is considered positive(+) ; Ct value >40isconsidered negative (-). For IC: Ct value ≤40 is considered positive (+); Ct value>40 is considered negative (-)
1. Reporting positive: Monkeypox Virus is detected.
2. Reporting negative: Monkeypox Virus is not detected. It is possible due tolowviral load and should be analyzed by combining clinical sign.
3. Invalid: Repeat sampling or collect specimen from different parts of the patient and repeat the test when clinical sign and other examinations are high suspected.
14. Performance Evaluation
14.1 Analytical Sensitivity
The limit of detection of the Bioperfectus Monkeypox Virus Real Time PCRKit forthe detection of DNA specific for Monkeypox virus from human tonsillar swab, nasopharyngeal swab, serum, whole blood, lesion exudate and scabwasdetermined to be 5 copies/reaction.
14.2 Analytical Specificity
No cross-reactivity of the Kit within the following selected microorganisms wereobserved.
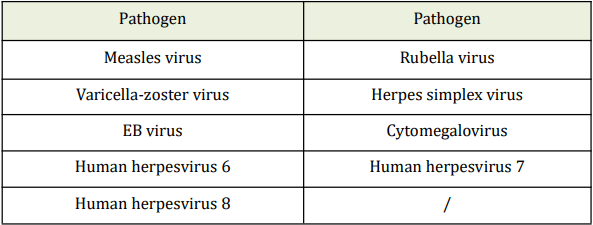
14.3 Precision
Precision references were used to evaluate the precision of BioperfectusMonkeypox Virus Real Time PCR Kit. The results show that, for the precisionreferences, coefficients of variation (CV%) of the repeatability andwithin-laboratory precision are less than 5%.
 Euro
Euro
 USD
USD
 British Pound
British Pound
 NULL
NULL